New tool predicts drug targets and IDs new anticancer compounds
Meyer Cancer Center researchers have created a new computational tool that could radically improve our ability to predict drug targets, identify novel anticancer compounds, find new applications for existing drugs, and expedite pharmaceutical research.
BANDIT - a Bayesian Approach to find Novel Drug Interaction Targets - mines millions of data points from five clinical, genomic, chemical and structural datasets to shed light on the targets and actions of certain molecules. Neel S. Madhukar, a graduate student in the lab of associate professor Olivier Elemento, Ph.D., presented the tool at the 2015 Annual Meeting of the American Association for Cancer Research in Philadelphia on April 21, along with other members of his research team: Linda Huang, Kaitlyn Gayvert and David Rickman.
“So far, no method has gone this far to predict, in such detail, a drug’s target, how it works, and its effect. The potential is enormous,” said Madhukar.
There are an estimated 20-30 million small molecules, including many natural products, but we only understand the workings of less than 0.01 percent. Identifying their targets has been a major challenge, driven by case specific experimentation - a slow and failure-prone process. Previous attempts to automate the process have relied on only a few types of data, leading to limited predictive capability.
Madhukar combined disjointed datasets—on drug efficacies, post-treatment transcriptional responses, drug structures, known adverse effects, and bioassay sensitivities from publicly-available databases—reconciling drug name differences along the way, an obstacle that had prevented other researchers from undertaking the task. He then spent several months culling and curating the databases, creating what is believed to be the largest and most comprehensive database of small molecule characteristics and their effects.
Madhukar fed the data into a program that computes how likely two molecules are to share a biological target. He first tested BANDIT using drug pairs known to share a target, and succeeded in predicting that target with 91 percent accuracy. After this proof of principle, BANDIT was used to predict new targets for known anticancer drugs, such as Vismodegib, currently used to inhibit the Hedgehog signaling pathway in basal cell carcinoma. BANDIT suggested its potential to also act as a tyrosine kinase inhibitor, which could be a valuable addition to the arsenal of anti-cancer agents.
Madhukar then applied BANDIT to the more than 50,000 compounds with unknown targets in the National Cancer Institutes (NCI) Developmental Therapeutics Program database to identify molecules that might be used for cancer therapy. He looked for unstudied drugs predicted to share a target with known anticancer drugs.
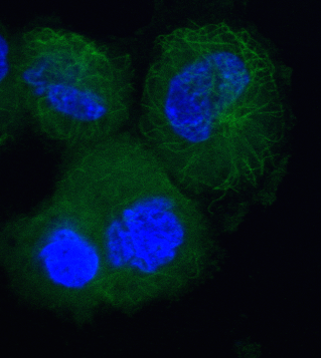
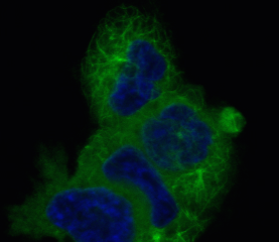
While seeking out potential taxane chemotherapy alternatives, BANDIT identified many new candidates, representing a range of structures and mechanisms of activity, pointing researchers down paths they might not have otherwise explored. Working with Paraskevi Giannakakou, associate professor of pharmacology in medicine, the team tested many of these candidates in an experimental setting and saw they did have significant effects on microtubule dynamics, as predicted.
“This not only serves as a validation of BANDIT's predictive capabilities but also opens the door for further testing to determine the therapeutic potential of these otherwise unknown candidates,” Madhukar said.
By identifying molecules that can target important regulatory proteins or are able to act upon multiple proteins along a pathway, the system might also provide an approach to targeting key kinases that are otherwise nearly impossible to treat. For newly discovered or created molecules, Elemento’s team can tell researchers which tests to run to generate the most useful data, which could then be fed through the program to determine likely targets.
“If you can’t figure out the target of a drug, it’s a failed drug. You can’t pursue it for clinical testing,” Elemento said. “This could significantly impact drug development and therapeutic decisions.”