Cantley team uncovers vulnerability that can be exploited to kill lung cancer cells
Researchers at Weill Cornell Medicine have gained insight into how alterations in a key metabolic pathway drive non-small cell lung cancers and lead to poor prognosis in its patients. In doing so, they may have also uncovered a potential new target for a cancer that is currently very difficult to treat.
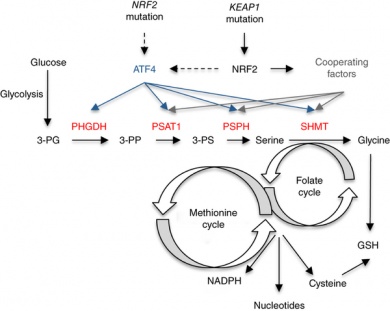
In a study published online Oct. 19 in the journal Nature Genetics, Gina DeNicola, Ph.D., and other members of the lab of Lewis Cantley, Ph.D., the Margaret and Herman Sokol Professor in Oncology Research at Weill Cornell Medicine and director of the Sandra and Edward Meyer Cancer Center, elucidated the role of Nrf2 in serine production in non-small cell lung cancer.
Nrf2 (nuclear factor erythroid-2-related like 2) is a transcription factor responsible for controlling proteins involved in cell defense and survival. It can protect cells and tissues from a variety of toxicants and carcinogens by increasing the expression of a number of cytoprotective genes. Studies have shown Nrf2 may also protect cancer cells from chemotherapeutic agents and facilitate cancer progression, leading to increased interest in understanding its role and its potential use in chemopreventive compounds.
Nrf2 is frequently deregulated in non-small cell lung cancer, and cancers with high Nrf2 levels are associated with poor prognosis, resistance to therapy, and rapid proliferation.
The Cantley Lab discovered that Nrf2 also plays a key role in the production of serine, which, together with glycine, provides the essential precursors for the synthesis of proteins, nucleic acids, and lipids that are crucial to cancer cell growth.
Crucial turning point
Cancer cells undergo specific metabolic reprogramming to sustain cell growth and proliferation. Glucose and glutamine are the main sources used to maintain the cancer cell’s anabolic metabolism, and the serine biosynthetic pathway represents a crucial turning point in glucose conversion. Some cells import serine from the blood stream, while others are able to make it. They convert the serine to glycine, which in turn provides carbon units for one-carbon metabolism. The synthesis of proteins, lipids, nucleic acids, and other cofactors requires one-carbon metabolism, which is a complex metabolite network that is based on the chemical reactions of folate compounds. Most current chemotherapies are antifolate treatments designed to attack this network.
Scientists believe that aberrant activation of the serine biosynthetic pathway is an essential process in cancer pathogenesis. They had already noticed that a key enzyme involved in the conversion of serine - phosphoglycerate dehydrogenase (PHGDH) – is upregulated in triple-negative breast cancer and in melanoma.
The Cantley Lab wondered if this was also the case in other cancers. Using a combination of metabolite tracing and gene expression analysis on large panels of cancer cell lines, the team uncovered and teased out the relationship between Nrf2, PHGDH regulation and activation of the serine biosynthetic pathway.
“We found the expression of the same enzyme responsible in breast cancer and melanoma, but the mechanism was completely different,” DeNicola said.
They tested their findings by silencing PHGDH in cancerous cells in cultures and tumor-ridden mice, and found that the cells did not grow or proliferate.
Taking target
Metabolic pathways can be difficult to attack with drugs. But because PHGDH is one of the first enzymes on the pathway, it could provide a viable target – and one that others in the Cantley Lab had already set their sights on.
“Potentially we could treat this with an inhibitor that targets the pathway,” DeNicola said. “This would be a big boost for non-small cell lung cancer, which typically has a poor prognosis. Patients tend to be resistant to traditional chemotherapy and radiation treatments, and so far nothing has really shown much promise.”
Their findings may also be applicable to other cancers where Nrf2 is overexpressed, such as pancreatic cancer, another disease that is currently very difficult to treat.
The team’s method for discovering the connection – by starting with a phenotype and tracing it back to the genome level by integrating metabolic tracing and transcriptional profiling – could also be more broadly applied to identify additional links between the activity of metabolic pathways and genetic alterations in cancers.
“It is a powerful tool for determining the mechanisms responsible for the differential regulation of metabolic pathways,” the authors wrote.
The work was funded in part by grants from the US. National Institutes of Health, the Welch Foundation, the Malcolm A.S. Moore Hope Funds for Cancer Research, and the PanCAN/AACR Pathway to Leadership.
“By disrupting this newly discovered cancer feeding mode, the work of Dr. DeNicola and her colleagues sets the stage for potential therapeutics to treat this deadly disease,” stated Frank Slack, Ph.D., Hope Funds Trustee and Scientific Advisor.
Co-authors include: Edouard Mullarkey and David Wu of Weill Cornell Medicine; Pei-Hsuan Chen, Jessica Sudderth, Zeping Hu, Hao Tang, Yang Xie, Kenneth Huffman, John D. Minna and Ralph J DeBerardinas of the University of Texas-Southwestern Medical Center; Ignacio Wistuba of MD Anderson Cancer Center; and John Asara of Harvard Medical School.