Driving discovery: Two teams uncover new player in prostate cancer formation
Their paths to discovery were completely different, but two teams of scientists at Weill Cornell Medicine arrived at the same place, and the result may impact on prostate cancer patients down the road.
For Ekta Khurana, Ph.D., assistant professor of computational genomics and member of the Sandra and Edward Meyer Cancer Center, the discovery of ERF as a driver of prostate cancer was proof that her new computational tool, RegNetDriver, is a valuable tool to elucidate the inner workings of tissue networks as they develop cancer.
For Juan Miguel Mosquera, M.D., M.Sc., associate professor of pathology and laboratory medicine, it highlighted the importance of including patient samples from a diverse population in research studies, as the anomaly was unearthed while interrogating tissue samples from African American prostate cancer patients.
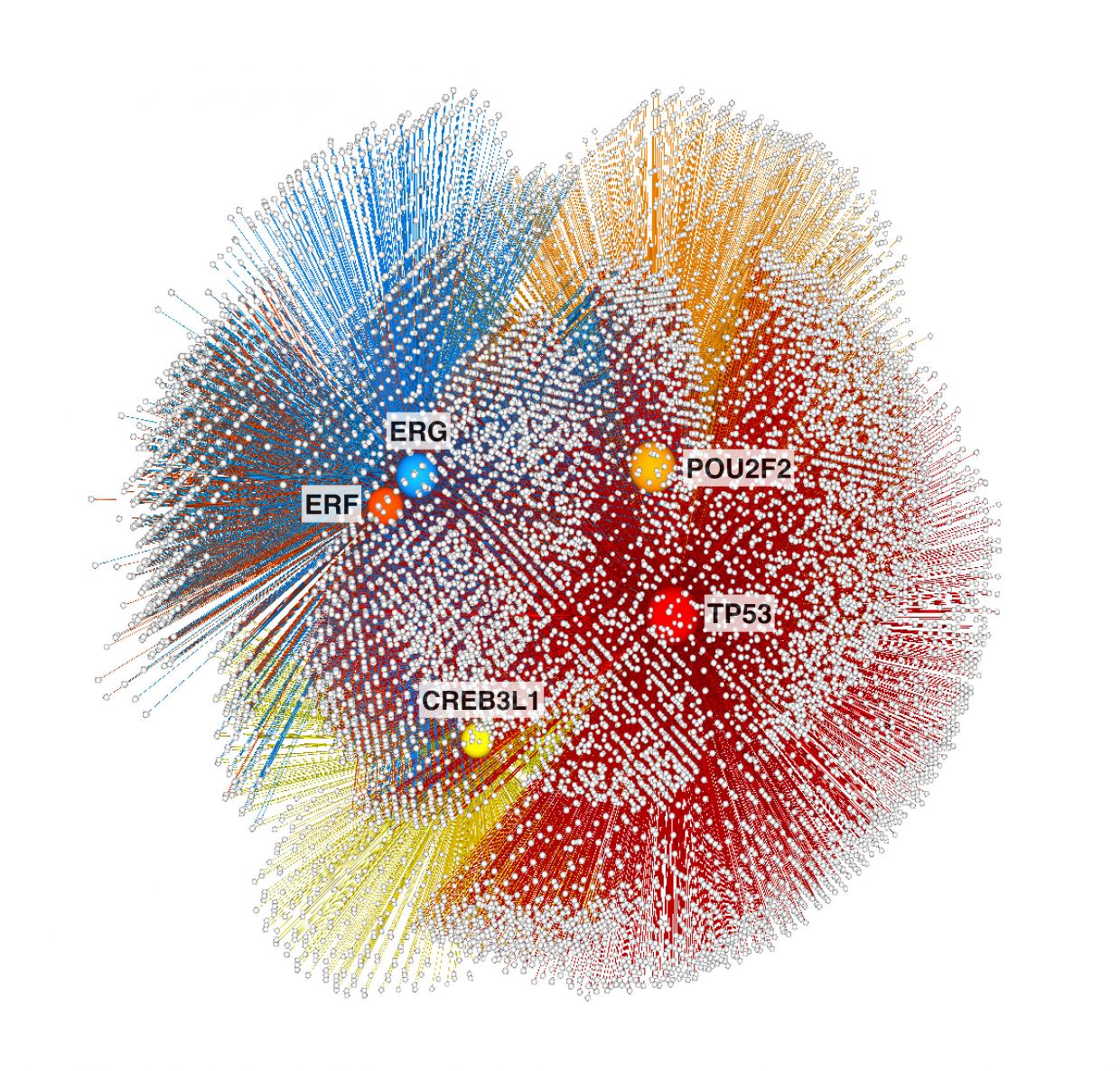
ERF is a “repressor” transcription factor, meaning it stops something from happening. Its analogue “activator” transcription factor is ERG. Both are members of the ETS transcription factor family, which is involved in a wide variety of functions including the regulation of cellular differentiation, cell cycle control, cell migration, cell proliferation, apoptosis (programmed cell death) and angiogenesis.
Altered expression of ERG due to gene fusions and deletions has already been implicated as a driver of prostate cancer, driving cancer cell growth out of control as if the organ’s gas pedal was stuck. What Khurana and Mosquera’s teams found were that altered expression of ERF due to mutations could also lead to speedy, out-of-control growth, as if its brakes were broken.
The purpose of Khurana’s RegNetDriver tool is to map out the role of each tiny part of the machinery of organs and tissue systems so that researchers and physician mechanics know where to look when things go wrong. Is the problem due to acceleration or failure to stop? Is it due to a worn brake pad, a lack of brake fluid, a leak, frayed line, bad alignment, a warped signal from the computer? A combination of these things? Just as each type of car is different, so is each type of tissue system. Some tissues are more likely to get brake (tumor suppressor) failure, while others are more likely to get stuck in hyperdrive (proto-oncogenes).
Knowing this, researchers can get a head start when forming their research plans and design their experiments using the right tools. Exome sequencing is useful when interrogating single nucleotide variants (SNVs), while whole genome sequencing is better at teasing out large structural variants (SVs) such as deletions, insertions, inversions, duplications and translocations.
In the case of prostate cancer, the first cancer that Khurana modeled using the regulatory network, as detailed in a July 27 study in Genome Biology, it seems that genomic alterations created by large SVs have a stronger regulatory impact than SNVs.
“So if you analyzed SNVs from exome sequencing of prostate cancer and didn’t find much, you are probably missing a lot,” Khurana said. “You have to know which types of events are important in which types of cancers.”
Khurana also included epigenetic alterations – changes caused by chemical modifications of DNA rather than the alteration of the genetic code itself – in her tissue network mapping. The team found that DNA methylation (a common signaling tool that can fix genes in the “off” position), can be impacted by the activity of powerful transcription factors that regulate a large number of genes and act as “hubs.”
“People have spoken of the phenomenon of transcription factor expression causing epigenetic changes.
This data supports that,” Khurana said.
By targeting transcription factor hubs, we might be able to reverse the cellular transformation from normal to malignant state and offer new therapeutic options, Khurana said. And if scientists are unable to target a repressor transcription factor such as ERF, it helps to know what its activator complement is (ERG), as it might be actionable.
It may sound simple, but was not. Mapping the flow of information from transcription factors to target genes via regulatory elements involved tracking more than a million unique interactions between 612 transcription factors and their 17,087 target genes. From that, Khurana’s group identified 153 transcription factor hubs that regulate the expression of thousands of downstream genes, including many with known connections to cancer. Khurana winnowed them down to five candidates most likely to be key cancer drivers – two (ERG and TP53) were well known, and three (ERF, CREB3LI and POU2F2) were new.
Independent validation
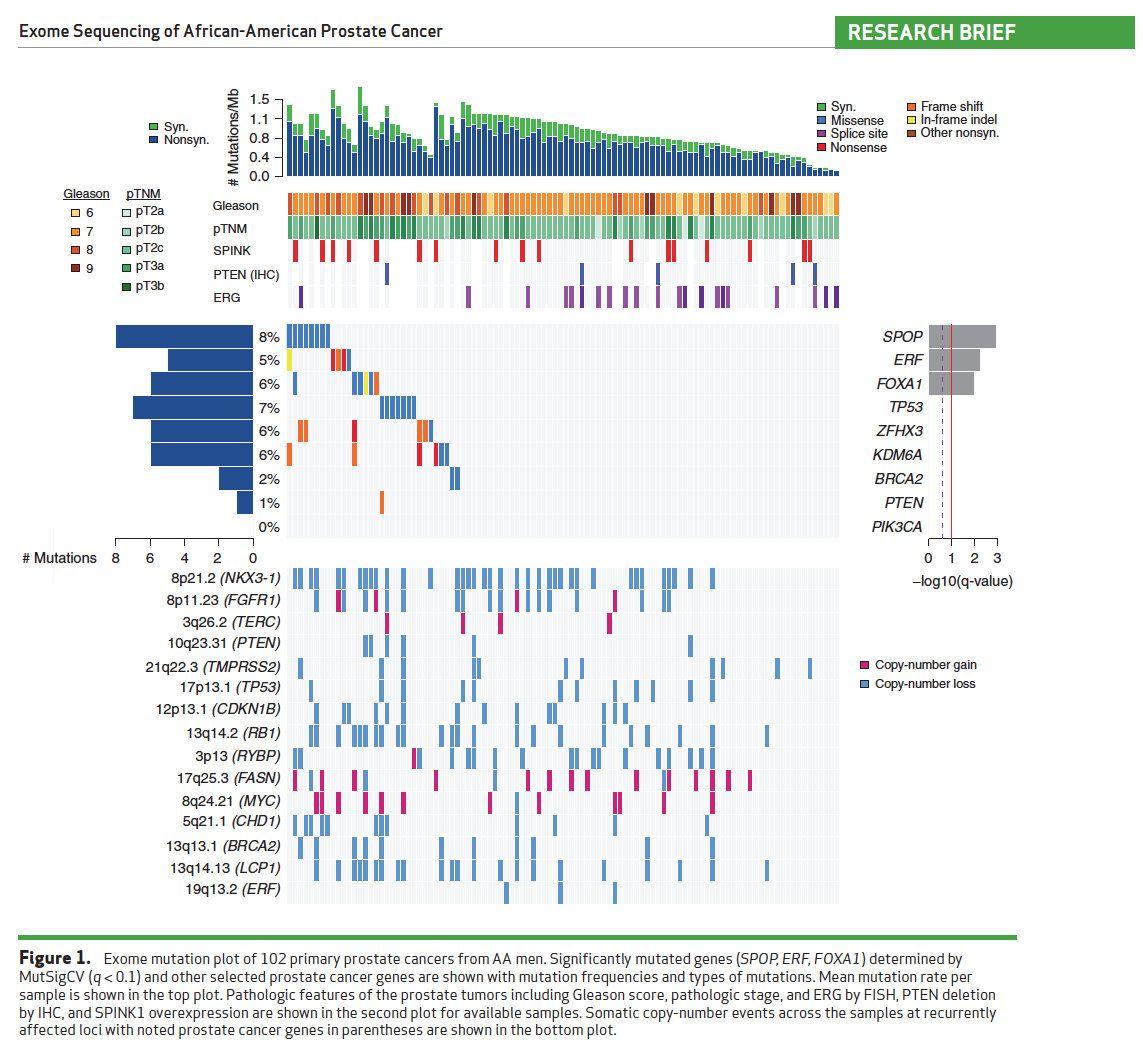
Meanwhile, the multi-institutional study published May 17 in Cancer Discovery, co-led by Drs. Mark A. Rubin and Juan Miguel Mosquera in collaboration with colleagues from the Broad Institute of MIT and Harvard University, also identified ERF as a tumor suppressor gene in prostate cancer. They came across the finding while analyzing sequencing data from 192 localized hormone-naïve prostate cancers in African-American men, which “identified several gene mutations not previously observed in this context.”
Among these was recurrent loss-of-function mutations in ERF, in 5 percent of cases. The team did further exploration of existing data from other sources and experiments in cell lines to validate the findings. Another paper from the Sawyers Lab at Memorial Sloan Kettering Cancer Center and the SU2C/PCF Prostate Cancer Dream Team, published July 14 in Nature, also explored the mechanics behind ERF and ERG activity in prostate oncogenesis.
"It’s very reassuring that using very different approaches, we got the same gene," Khurana said.
In addition to ERF mutations, the Weill Cornell and Broad teams found other differences in mutational events in African-American prostate cancers. For example, one mutation (PTEN deletions) found in 32 percent of the mostly European cohort represented in the Cancer Genome Atlas (TCGA) dataset, was found in only 6 percent of the 192 African-American men sequenced for the Mosquera et al. study, whereas another set of mutations within the fatty acid synthase network occurred in 12.7 percent of the African-Americans and only 1 percent of the TCGA cohort.
“African-American men, who have a 1.4-fold higher incidence and 2.4-fold higher mortality rate from prostate cancer compared to non-Hispanic whites, have been underrepresented in most systematic studies of prostate cancer performed to date. Of the 333 primary prostate cancer profiles in the TCGA, for example, 270 (81 percent) are from men of European ancestry,” Rubin, co-senior author of the study said.
“Our results indicate that inclusion of sufficient numbers of patients of African ancestry in cancer genomics studies may enable the discovery of new cancer genes and inform the inclusion of diverse populations towards precision cancer medicine,” added Mosquera, who also leads the Pathology Team at the Caryl and Israel Englander Institute for Precision Medicine.
The work is also relevant to other cancers. Mutations in ERF occur in other tumor types, including stomach and colon cancers. And Khurana’s tool can be used to model other tissue networks, to create regulatory guides for a variety of tumor types.